Mass photometry analysis of low-affinity protein interactions
Mass photometry & low-affinity interactions
Mass photometry is a valuable bioanalytic technique for rapidly measuring the mass distribution of biomolecules in solution, without the need for labels. Typically, mass photometry works optimally at sample concentrations below 100 nM. While this concentration is suitable for many measurement contexts, in some cases it is necessary to analyze sample behavior at a higher concentration. For example, complexes formed through low-affinity interactions dissociate at low protein concentrations, meaning they can often be reliably detected only at higher concentrations.

How does MassFluidix HC expand the concentration range for mass photometry measurements?
MassFluidix HC is a microfluidics system that significantly broadens the range of sample concentrations amenable to mass photometry analysis. It is an add-on that allows mass photometry measurements at micromolar sample concentrations.
The MassFluidix HC system enables a measurement to be made at a concentration that is optimal for mass photometry, while capturing the state of the biomolecular interactions at micromolar concentration.
The system works by rapidly diluting the sample and flowing it across the measurement surface very quickly – before the biomolecular interaction equilibrium has been disrupted by the dilution.
Under these conditions, the complex will remain intact when the sample is measured.
MassFluidix HC helps reveal low-affinity complexes
The MassFluidix HC system add-on can help reveal low-affinity complexes that were previously undetected. In the example provided in the application note, the MassFluidix HC system was used with the TwoMP mass photometer – to investigate the oligomerization behavior of integrin αL/β2, a low-affinity integrin heterodimer.
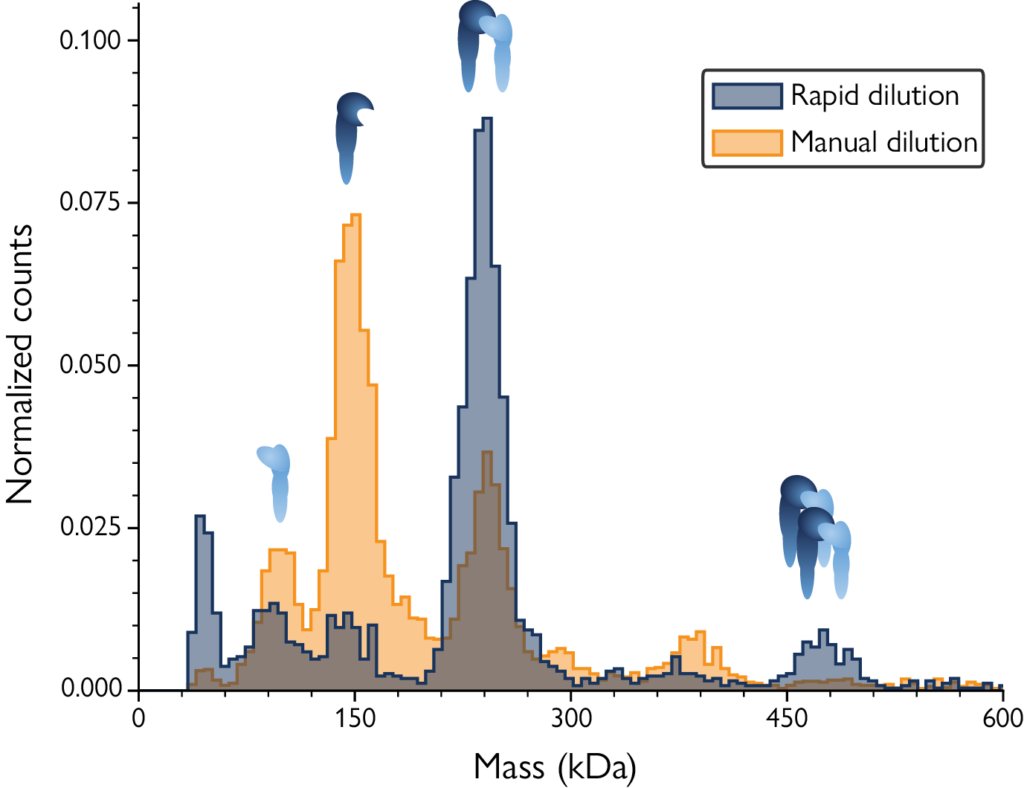
After manual dilution, the mass profile showed mainly monomers of αL and β2 with a small peak corresponding to heterodimers. After rapid dilution with MassFluidix HC, the sample state at micromolar concentration was captured, showing the presence of more dimers and bigger complexes with a corresponding decrease in monomers – revealing low-affinity interactions that occur at high sample concentrations. For more details about this study, read the application note.
Image: Mass histograms reveal low-affinity complexes only after rapid sample dilution with MassFluidix HC. For experimental details, download the app note.
To learn how mass photometry can help characterize low-affinity interactions
Mass photometry analysis of samples at micromolar concentration
Learn how mass photometry facilitates the analysis of biomolecular interactions, enabling the characterization of protein binding, oligomerization and macromolecular assembly –all in a label-free, solution-based approach. Find out how you can significantly broaden the sample concentration range for your mass photometry analysis with the mass photometry add-on MassFluidix HC. Discover how the system’s rapid dilution function allows you to expand the range of sample concentrations you can analyze with mass photometry, ultimately enabling you to explore and characterize low-affinity interactions that were previously challenging to study.
Additional resources
APPLICATION NOTE: Characterization of protein interaction equilibria
Learn how mass photometry can be applied to the study of complex equilibrium formation and assess how changes in the chemical environment or protein concentration affect the equilibria. More specifically, discover how mass photometry was used to study the oligomerization process of a recombinant protein that forms tetramers. Using this multimeric protein as an example, you can see how scientists measured the relative abundances of each oligomeric species across a range of physiologically relevant protein concentrations and assessed the effect of environmental factors (such as temperature) on reaction equilibria.

APPLICATION NOTE: Quantifying protein binding affinities using mass photometry
Learn how mass photometry can be used to assess the dynamics and strength of protein-protein interactions. Discover how it can be used to characterize interactions between Immunoglobulin G (IgG) antibodies of different origin species (human and bovine) and protein A, by quantifying the relative abundance of each protein and the complexes they form in solution. Gain insights on how the automated pipetting feature of Refeyn’s TwoMP Auto mass photometer generates highly reproducible data. Finally, learn how you can calculate the equilibrium dissociation constant (KD) for each protein interaction from mass photometry data.
To discuss how mass photometry can be applied in protein sample analysis at micromolar concentrations
More Application Notes
Browse through our catalogue of application notes highlighting some recent case studies featuring mass photometry.
