Analysis of protein-protein interaction equilibria
Studying protein-protein interactions in solution
Understanding protein-protein interactions is crucial for deciphering the intricacies of cellular processes, as these interactions play pivotal roles in diverse functions, ranging from DNA replication to cell cycle control. Characterizing protein interactions provides insights into reaction kinetics and binding affinity, deepening our grasp of molecular mechanisms driving cellular activities.
How to study protein reaction equilibria?
In the study of reaction equilibria, mass photometry can be used to examine how protein-protein interaction systems regain equilibrium after perturbations. For instance, when investigating a recombinant protein behavior exposed to stress at physiologically relevant concentrations, mass photometry can reveal changes in sample composition and show how populations of monomers, dimers, and tetramers form, shedding light on equilibrium restoration.
How to study protein reaction equilibria?
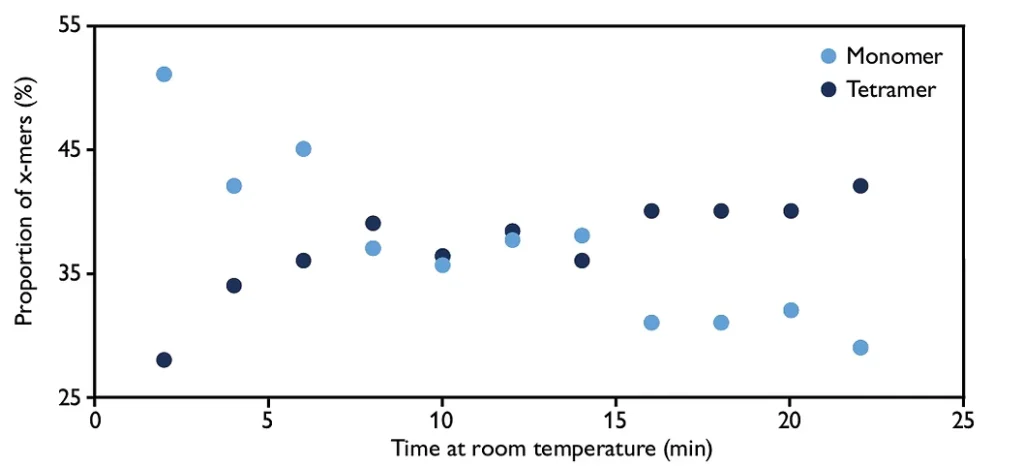
Environmental factors like temperature, ionic strength, and pH can significantly impact protein-protein interaction equilibria. Altering the temperature or chemical environment can perturb the dynamic equilibrium, resulting in changes in the relative proportions of various protein species.
Mass photometry single-molecule approach enables rapid and accurate measurements, making it an invaluable tool for verifying the state of a protein system of interest and gaining insights into the effects of environmental factors on protein-protein interactions.
Image: Mass photometry was used to assess the effect of temperature on protein oligomerization. For experimental details, download the app note
To learn how to characterize protein-protein interactions with mass photometry
Mass photometry of protein interaction equilibria
Learn how mass photometry can be applied to the study of complex equilibrium formation and assess how changes in the chemical environment or protein concentration affect the equilibria. More specifically, discover how mass photometry was used to study the oligomerization process of a recombinant protein that forms tetramers. Using this multimeric protein as an example, you can see how scientists measured the relative abundances of each oligomeric species across a range of physiologically relevant protein concentrations and assessed the effect of environmental factors (such as temperature) on reaction equilibria.
Additional resources
APPLICATION NOTE: Quantifying protein binding affinities using mass photometry
Learn how mass photometry can be used to assess the dynamics and strength of protein-protein interactions. Discover how it can be used to characterize interactions between Immunoglobulin G (IgG) antibodies of different origin species (human and bovine) and protein A, by quantifying the relative abundance of each protein and the complexes they form in solution. Gain insights on how the automated pipetting feature of Refeyn’s TwoMP Auto mass photometer generates highly reproducible data. Finally, learn how you can calculate the equilibrium dissociation constant (KD) for each protein interaction from mass photometry data.


WEBINAR: Monitoring aggregation levels of biosimilar mAbs using mass photometry and SEC
Learn how mass photometry compares with size-exclusion chromatography in measuring aggregation levels of monoclonal antibodies. Discover the pros and cons of each technology and the factors that should be considered when analyzing and comparing data generated by these different techniques.
To discuss how mass photometry can be applied to the characterization of protein interaction equilibria
More Application Notes
Browse through our catalogue of application notes highlighting some recent case studies featuring mass photometry.
