Enabling rapid structural studies
Mass photometry is compatible with most membrane mimetics and offers a non-destructive method for optimizing extraction and purification conditions – enabling structural studies of transmembrane proteins by cryo-EM, X-ray crystallography and other methods.
Mass photometry supports membrane protein analysis
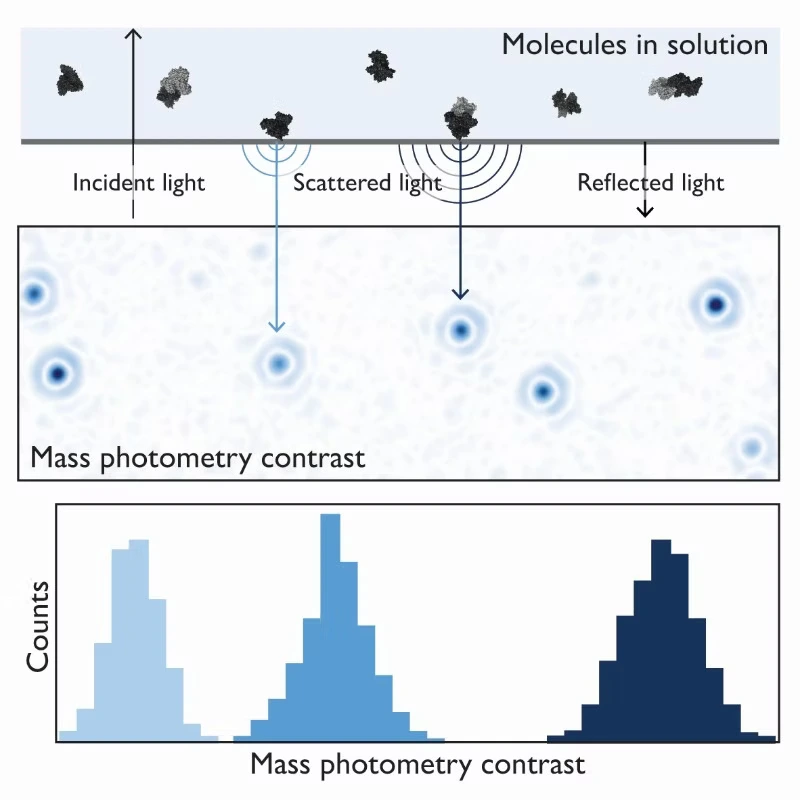
Mass photometry is a powerful analytical technique that can help overcome the bottlenecks associated with membrane protein characterization. It quantifies samples at the single-molecule level by measuring the light scattered by individual biomolecules in solution (Figure 1). Its versatility extends to a wide array of applications, from assessing assembled/disassembled complexes to identifying impurities and protein aggregates.
Mass photometry is uniquely equipped to deliver fast, precise results using minimal sample input, and is compatible with most membrane mimetics. As this process is non-destructive and does not require labels, researchers can identify optimal conditions for their structural studies without wasting valuable resources. Furthermore, its user-friendly nature and small instrument footprint make mass photometry highly accessible and adaptable for routine use in laboratory settings.
Figure 1. The principle of mass photometry. The light scattered by a molecule that has landed on a measurement surface interferes with light reflected by that surface. The interference signal scales linearly with mass.
Compatibility with detergents
Detergents are a popular method for membrane protein purification because of their comparative ease of use. However, detergents may destabilize and denature proteins, in addition to increasing sample heterogeneity. Given the huge variety of detergents currently available, identifying a detergent that fulfils specific protein requirements, in terms of stability and functionality, can be a painstaking trial-and-error process.
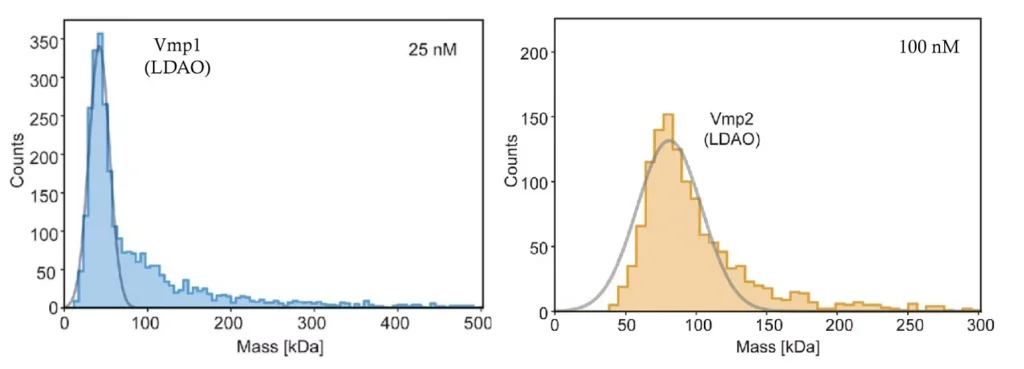
Mass photometry not only eliminates the necessity for complete detergent removal but also enables quick assessment of solubility conditions in a wide range of buffers (Figure 2). As measurements take less than five minutes, researchers can rapidly analyze different detergent concentrations and buffer conditions to pinpoint the optimal parameters for membrane protein stability and functionality.
Compatibility with other membrane mimetics
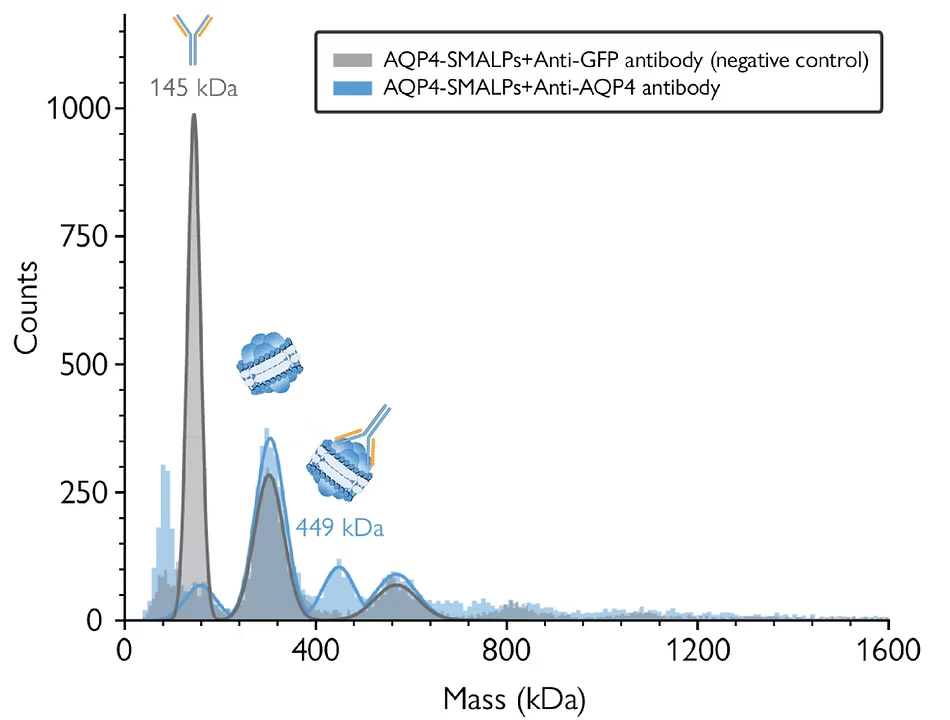
Mass photometry is readily compatible with a variety of membrane mimetics beyond detergents, including nanodiscs, SMALPs, and peptidiscs (Young et al., 2024). The high resolution of mass photometry enables the analysis of complex samples, including nanodiscs of varying compositions or sizes. Operating under non-disruptive conditions, mass photometry enables measurements of samples embedded within nanodiscs, facilitating the assessment of results from antibody assays or functional assays (Figure 3), as showcased in studies like Olerinyova et al. (2021). This capability provides valuable insights into the structure and function of membrane proteins.
Mass photometry solutions for protein interaction analytics
- TwoMP
-

For molecular mass measurements with unmatched sensitivity, speed and simplicity of use, a TwoMP mass photometer offers a wide mass range and single-molecule resolution.
High-fidelity measurements of molecular mass
Little sample required
Intuitive acquisition and data analysis software
Easy setup – a compact, benchtop instrument with minimal installation requirements
- TwoMP Auto
-

The TwoMP automated mass photometer frees up operator time and offers enhanced precision, enabling rapid measurement of multiple samples with low sample consumption.
Rapid, automated measurement of multiple samples
Ideal for screening and titration assays
Highly reproducible data with user-friendly software
- Refeyn consumables
-
 By using Refeyn consumables, you can spend less time preparing for measurements and gain greater confidence in your data. The consumables range includes calibrants, sample carrier slides and more.
By using Refeyn consumables, you can spend less time preparing for measurements and gain greater confidence in your data. The consumables range includes calibrants, sample carrier slides and more.

For molecular mass measurements with unmatched sensitivity, speed and simplicity of use, a TwoMP mass photometer offers a wide mass range and single-molecule resolution.
High-fidelity measurements of molecular mass
Little sample required
Intuitive acquisition and data analysis software
Easy setup – a compact, benchtop instrument with minimal installation requirements

The TwoMP automated mass photometer frees up operator time and offers enhanced precision, enabling rapid measurement of multiple samples with low sample consumption.
Rapid, automated measurement of multiple samples
Ideal for screening and titration assays
Highly reproducible data with user-friendly software
 By using Refeyn consumables, you can spend less time preparing for measurements and gain greater confidence in your data. The consumables range includes calibrants, sample carrier slides and more.
By using Refeyn consumables, you can spend less time preparing for measurements and gain greater confidence in your data. The consumables range includes calibrants, sample carrier slides and more. What mass photometry users are saying
Saponaro et al. (2022) Front. Physiol.,
Philip Kitchen
School of Biosciences, Aston University
View further resources related to membrane protein analytics
- Selected publications
-
Niebling, S., Veith, K., Vollmer, B. et al. 2022. Biophysical screening pipeline for Cryo-EM grid preparation of membrane proteins. Frontiers in Molecular Biosciences, 9, p.882288. [Link]
Young, J.W., Pfitzner, E., van Wee, R., et al. 2024. Characterization of membrane protein interactions by peptidisc-mediated mass photometry. Iscience, 27(2). [Link]
Olerinyova, A., Sonn-Segev, A., Gault, J., Eichmann, C., et al. 2021. Mass photometry of membrane proteins. Chem, 7(1), pp.224-236. [Link]
Weiland, P. and Altegoer, F., 2021. Identification and characterization of two transmembrane proteins required for virulence of Ustilago maydis. Frontiers in Plant Science, 12, p.669835. [Link]
Webby, M.N., Oluwole, A.O., Pedebos, C., Inns, P.G., Olerinyova, A., Prakaash, D., Housden, N.G., Benn, G., Sun, D., Hoogenboom, B.W. and Kukura, P., (2022). Lipids mediate supramolecular outer membrane protein assembly in bacteria. Science Advances, 8(44), p.eadc9566. [Link]
- Vostal, L.E., 2025. Need for speed: Mass photometry as a sample analysis tool for structural studies. Structure, 33(6), pp.994-996. [Link]
- Webinars
-
- App notes and white papers
-
- Refeyn posts
-
- Brochure
-
- External articles
-
Niebling, S., Veith, K., Vollmer, B. et al. 2022. Biophysical screening pipeline for Cryo-EM grid preparation of membrane proteins. Frontiers in Molecular Biosciences, 9, p.882288. [Link]
Young, J.W., Pfitzner, E., van Wee, R., et al. 2024. Characterization of membrane protein interactions by peptidisc-mediated mass photometry. Iscience, 27(2). [Link]
Olerinyova, A., Sonn-Segev, A., Gault, J., Eichmann, C., et al. 2021. Mass photometry of membrane proteins. Chem, 7(1), pp.224-236. [Link]
Weiland, P. and Altegoer, F., 2021. Identification and characterization of two transmembrane proteins required for virulence of Ustilago maydis. Frontiers in Plant Science, 12, p.669835. [Link]
Webby, M.N., Oluwole, A.O., Pedebos, C., Inns, P.G., Olerinyova, A., Prakaash, D., Housden, N.G., Benn, G., Sun, D., Hoogenboom, B.W. and Kukura, P., (2022). Lipids mediate supramolecular outer membrane protein assembly in bacteria. Science Advances, 8(44), p.eadc9566. [Link]
- Vostal, L.E., 2025. Need for speed: Mass photometry as a sample analysis tool for structural studies. Structure, 33(6), pp.994-996. [Link]
