Analyzing protein-protein interactions with mass photometry
Analyzing protein-protein interactions
Protein-protein interactions underpin virtually all cellular processes. Studying these interactions can be challenging however, since they often involve multiple components and complex, dynamic equilibria. Many of the resulting protein complexes appear at very low concentrations, making them difficult to detect and quantify with traditional analytical methods.
Mass photometry is a powerful solution for analyzing protein-protein interactions. It rapidly provides detailed information on the mass distribution of protein samples at a single-molecule level, enabling the detection and quantification of trace protein complexes and transient interactions.
On this Page
- A powerful technique for protein analytics
- Gain quantitative insights into protein-protein interactions
- Investigating weak interactions with MassFluidix HC
- Characterizing protein oligomerization with mass photometry
- Mass photometry solutions for protein interaction analysis
- What mass photometry users are saying
- Additional resources
A powerful technique for protein analytics
Whether investigating the assembly of large, multimeric complexes or probing oligomerization behavior, mass photometry delivers rapid and accurate insights into the full range of species present in a sample.
By measuring the mass of individual proteins and complexes in a sample, mass photometry quantifies the relative abundance of each species present in a protein-protein interaction at equilibrium. This enables users to identify the complexes formed, assess their stoichiometry, and evaluate the strength of the interactions by determining the dissociation constant (KD). The method is label-free and non-destructive – helping you to achieve optimal reproducibility in your results.
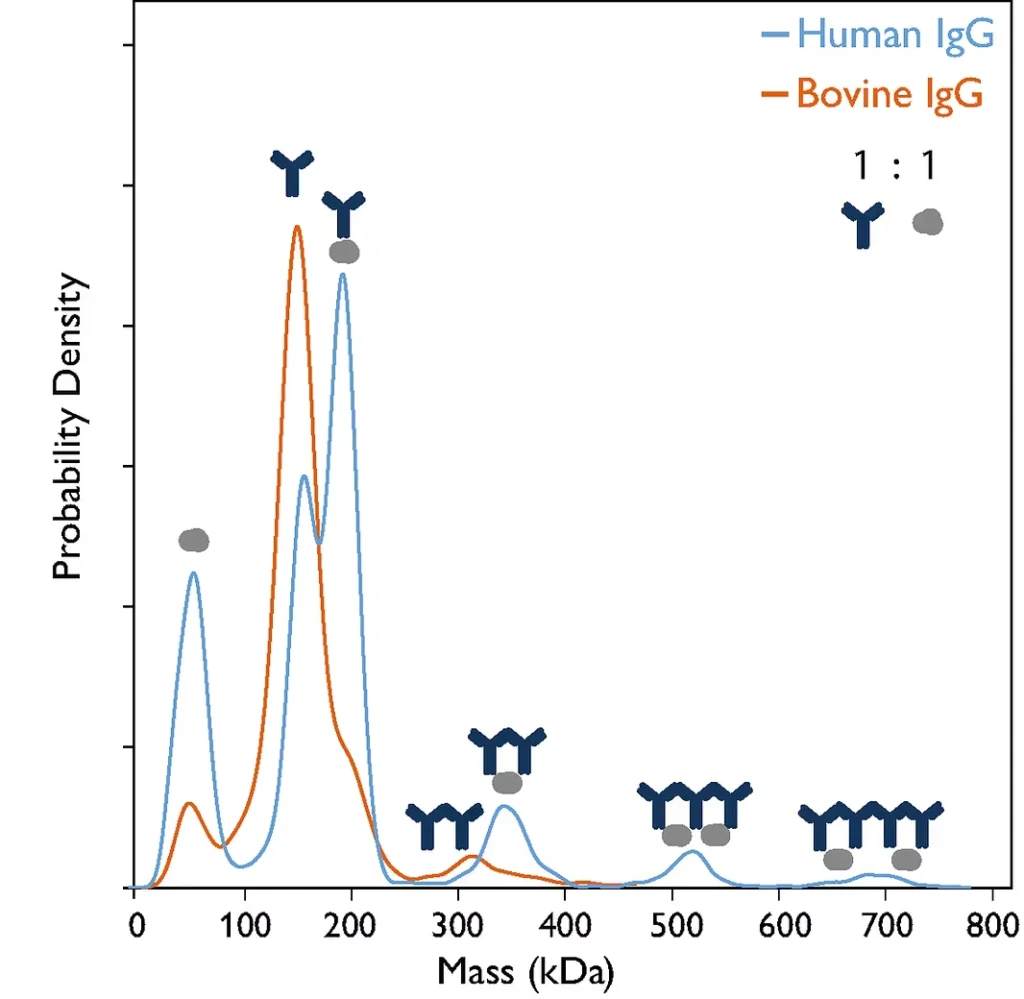
Figure 1: Mass photometry reveals differences in complex formation between protein A and IgG antibodies of differing origin. When human IgG was mixed with protein A at a 1:1 ratio (blue line), several species could be resolved, including Protein A; IgG; and protein A: IgG complexes of stoichiometry ratios 1:1, 1:2, 2:3 and 2:4. When bovine IgG was mixed with protein A (orange line), Protein A, IgG, 1:1 protein A: IgG complex and IgG dimers could be resolved.
Gain quantitative insights into protein-protein interactions
The value of mass photometry for protein-protein interaction analysis lies in the rapid quantitative overview it provides of the biomolecular species and complexes present in a sample. In addition, its capability to perform measurements across a diverse array of buffer conditions makes the technology well-suited for evaluating protein interactions under a variety of physiologically relevant conditions.
The TwoMP mass photometer delivers protein analytics results in minutes, enabling researchers to observe the evolution of protein interactions over time, and therefore characterize reaction equilibria and dynamics efficiently and effectively.
A major advantage of mass photometry is its single-molecule resolution, which affords precise quantification of the relative amounts of molecular species in a sample. This is especially valuable when analyzing protein components present at low concentrations.
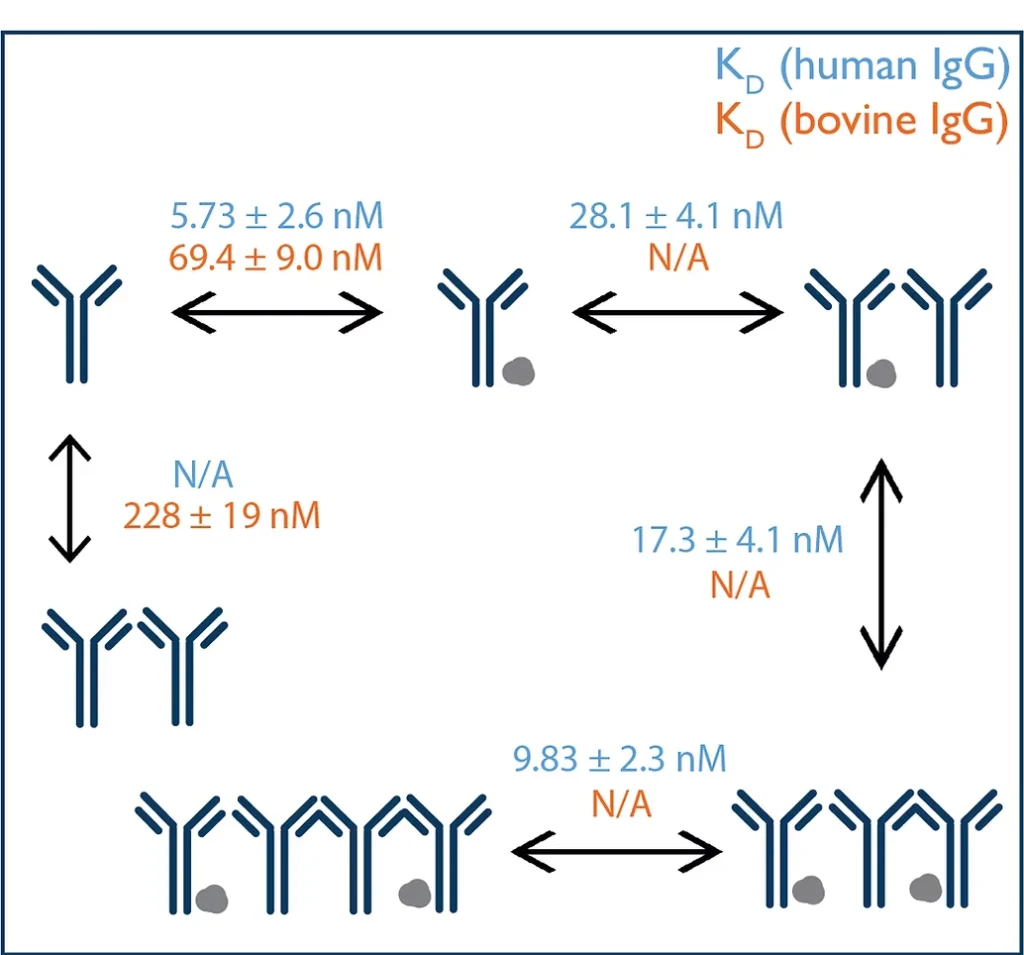
Figure 2: KD values for interactions involving the IgG of human origin (blue) and of bovine origin (orange) are both shown. The KD calculation was not applicable for certain interactions (depicted as N/A). KD values were calculated for each of three repeated mass photometry measurements, and the mean ± standard deviation is given.
Investigating weak interactions with MassFluidix HC
MassFluidix High Concentration (HC) is a microfluidics system that works in conjunction with the TwoMP mass photometer to expand the sample concentration range available for mass photometry analysis. The nanomolar sample concentrations needed for mass photometry can be below the threshold for studying very weak or transient interactions that naturally occur at higher concentrations. MassFluidix HC harnesses microfluidics to rapidly dilute samples immediately prior to measurement, allowing input sample concentrations up to the tens of micromolar range.
The increased sample concentrations that can be analyzed using MassFluidix HC enable researchers to explore a wider array of protein behaviors in physiologically relevant conditions, capturing interactions that play crucial roles in biological processes. The microfluidic device can also provide deeper insights into the concentration-dependent behavior of protein complexes, allowing for more precise characterization of interaction strengths and dynamics.
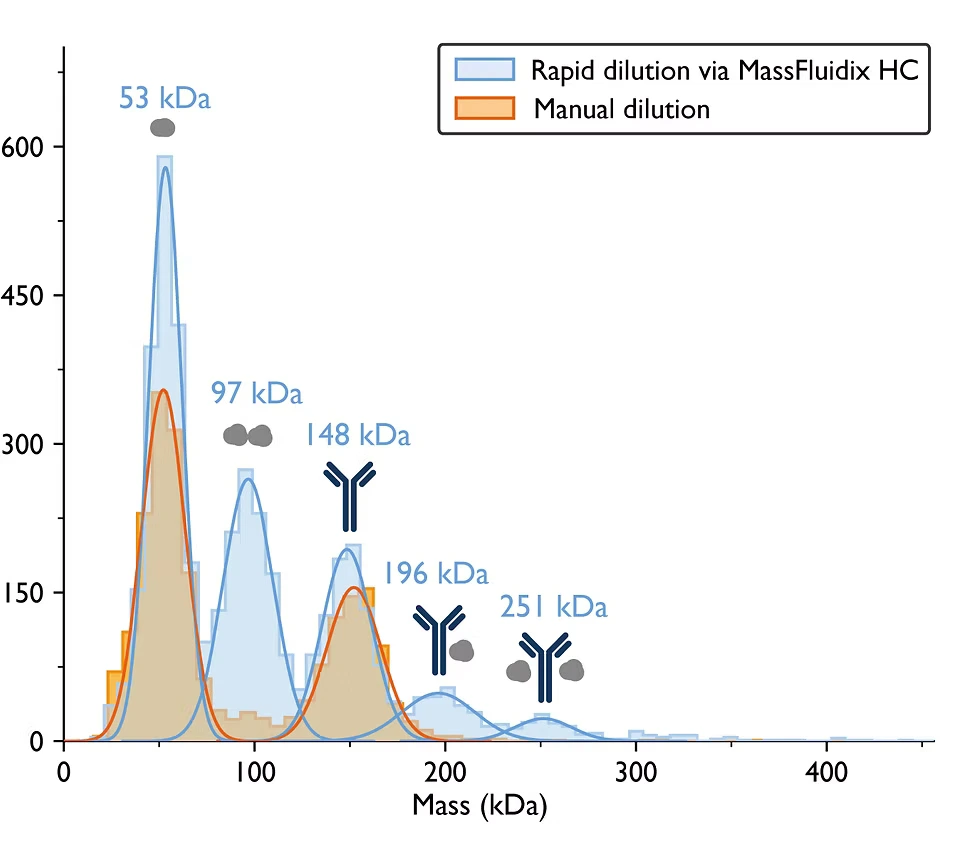
Figure 3. Rapid dilution by MassFluidix HC reveals low-affinity complexes. The IgG antibody was mixed with the soluble domain of the IgG neonatal Fc receptor (FcRn) at a 1:10 ratio (pH 5.0). After manual dilution (shown in orange), peaks corresponding to FcRn monomers and IgG monomers were detected. After rapid dilution with MassFluidix HC (shown in blue), additional peaks corresponding to FcRn dimers and IgG-FcRn complexes with 1:1 and 1:2 stoichiometry were also observed.
Characterizing protein oligomerization with mass photometry
Oligomerization, the self-assembly of proteins into specific quaternary structures, plays a vital role in protein functionality. This self-assembly is essential for numerous proteins to perform their biological roles effectively. For example, oligomerization states can regulate the activity of enzymes by allosteric inhibition.
Mass photometry characterizes these oligomeric behaviors with high precision. It can detect species that represent less than 1% of the molecules in a sample, providing insights into the diversity of oligomeric states and their role in protein function. This high-resolution capability ensures that you can identify and analyze even the most transient and low-abundance complexes, offering a comprehensive view of protein behaviors.
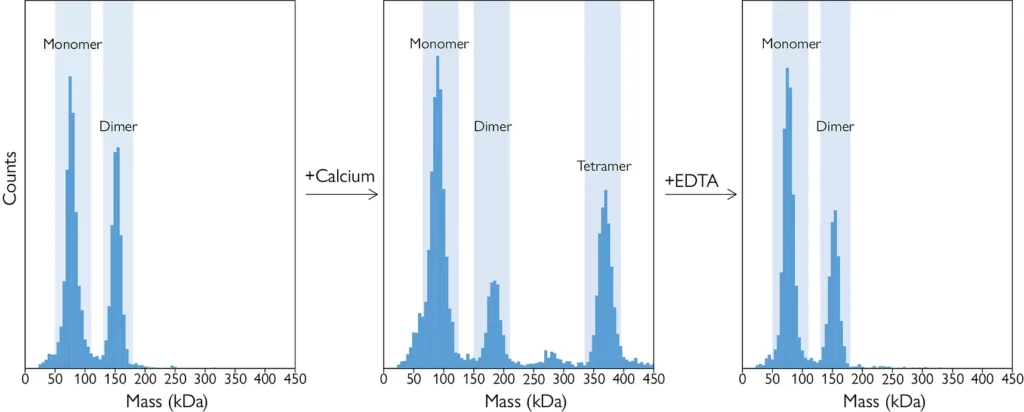
Figure 3: A mass histogram of a protein-only sample, with monomers and dimers present. B: Calcium chloride (present in excess) induced the formation of tetramers and reduced the number of dimers. C: Adding a saturating concentration of EDTA reversed this effect, resulting in the disappearance of the tetrameric species.
Delivering results swiftly, mass photometry is an ideal tool for screening protein variants for oligomerization behavior. With the TwoMP mass photometer, you can simultaneously evaluate protein oligomer variants under various conditions, revealing how environmental factors influence protein assembly. Since the concentrations used in mass photometry experiments are within the nanomolar range, similar to physiological conditions, the observed oligomeric states closely mirror the protein’s behavior in vivo.
Further enhancing its utility in protein oligomerization studies, the TwoMP Auto mass photometer can automate oligomerization analysis, allowing for repeated measurements and hands-free analysis of up to 24 samples. The addition of the MassFluidix HC microfluidics add-on supercharges the capability of the TwoMP mass photometer to characterize low-affinity interactions. Harnessing microfluidics to rapidly dilute samples prior to measurement, it uncovers complexes that would remain undetected at the nanomolar concentrations typically used in mass photometry. This opens novel avenues for the exploration of transient and weak interactions, offering a fuller picture of protein behavior in physiological and experimental conditions.
Mass photometry solutions for protein interaction analytics
- TwoMP
-

For molecular mass measurements with unmatched sensitivity, speed and simplicity of use, a TwoMP mass photometer offers a wide mass range and single-molecule resolution.
High-fidelity measurements of molecular mass
Little sample required
Intuitive acquisition and data analysis software
Easy setup – a compact, benchtop instrument with minimal installation requirements
- TwoMP Auto
-

The automated mass photometer frees up operator time and offers enhanced precision, enabling rapid measurement of multiple samples with low sample consumption.
Rapid, automated measurement of multiple samples
Ideal for screening and titration assays
Highly reproducible data with user-friendly software
- MassFluidix HC
-

Refeyn’s microfluidics system, MassFluidix HC, significantly expands the range of sample concentrations amenable to investigation by mass photometry, by raising the upper sample concentration limit from the nanomolar to the micromolar range. This enables applications such as the characterization of low-affinity interactions.
- Enables measurement of concentrated samples
- Uses rapid dilution
- Is an add-on for TwoMP and OneMP mass photometers
- Refeyn consumables
-
 By using Refeyn consumables, you can spend less time preparing for measurements and gain greater confidence in your data. The consumables range includes calibrants, sample carrier slides and more.
By using Refeyn consumables, you can spend less time preparing for measurements and gain greater confidence in your data. The consumables range includes calibrants, sample carrier slides and more.

For molecular mass measurements with unmatched sensitivity, speed and simplicity of use, a TwoMP mass photometer offers a wide mass range and single-molecule resolution.
High-fidelity measurements of molecular mass
Little sample required
Intuitive acquisition and data analysis software
Easy setup – a compact, benchtop instrument with minimal installation requirements

The automated mass photometer frees up operator time and offers enhanced precision, enabling rapid measurement of multiple samples with low sample consumption.
Rapid, automated measurement of multiple samples
Ideal for screening and titration assays
Highly reproducible data with user-friendly software

Refeyn’s microfluidics system, MassFluidix HC, significantly expands the range of sample concentrations amenable to investigation by mass photometry, by raising the upper sample concentration limit from the nanomolar to the micromolar range. This enables applications such as the characterization of low-affinity interactions.
- Enables measurement of concentrated samples
- Uses rapid dilution
- Is an add-on for TwoMP and OneMP mass photometers

Testimonials from mass photometry users
John Zinder
Senior scientist at Odyssey Therapeutics
“The biggest advantage of mass photometry is that we could assay so many conditions in a high throughput way. We were able to characterize the monomer-to-dimer transition in a low concentration range, which would have been difficult to detect with other techniques.”
Kathryn Gunn
Postdoctoral Fellow, The University of North Carolina at Chapel Hill
“I will try to keep mass photometry as integrated in my work as possible because it is a very honest technique that gives you a direct picture of your sample and there is not a lot of room for interpretation.”
Luca Schulz
graduate student at the Max Planck Institute for Terrestrial Microbiology in Marburg
“Previous gel filtration chromatography experiments indicated that one of the proteins I studied was a hexamer… I subsequently did the same experiment using mass photometry and I realized that my protein exists in various oligomeric forms… This revelation would not be possible without mass photometry!”
Ananya Acharya
Recombination Mechanisms PhD Student, USI Switzerland
Wu and Piszczec (2020)
“The low sample and time requirements of mass photometry make it valuable to us. We use it mainly to determine the stoichiometry of protein complexes and to check if they are adequately formed before doing structural analysis. It turned out to be a very good machine!”
Kai-En Chen
postdoctoral researcher at the Institute for Molecular Bioscience (IMB), University of Queensland (Brisbane, Australia)
View further resources related to protein-protein interactions analytics
- Selected publications
-
Kofinova, Z., Karunanithy, G., Ferreira, A.S. and Struwe, W.B., 2024. Measuring protein‐protein interactions and quantifying their dissociation constants with mass photometry. Current Protocols, 4(1), p.e962 [Link]
Claasen, M., Kofinova, Z., Contino, M. and Struwe, W.B., 2024. Analysis of protein complex formation at micromolar concentrations by coupling microfluidics with mass Photometry. Journal of Visualized Experiments: Jove, (203). [Link]
Nagy, G.N., Zhao, X.F., Karlsson, R., Wang, K., Duman, R., Harlos, K., El Omari, K., Wagner, A., Clausen, H., Miller, R.L. and Giger, R.J., 2024. Structure and function of Semaphorin-5A glycosaminoglycan interactions. Nature Communications, 15(1), p.2723. [Link]
Awasthi, S., Ying, C., Li, J. and Mayer, M., 2023. Simultaneous determination of the size and shape of single α-synuclein oligomers in solution. ACS nano, 17(13), pp.12325-12335. [Link]
Wu, D. and Piszczek, G., 2020. Measuring the affinity of protein-protein interactions on a single-molecule level by mass photometry. Analytical biochemistry, 592, p.113575. [Link]
- Webinars
-
- Tau protein aggregation & neurodegenerative disease; new insights from mass photometry
- Characterizing protein oligomerization dynamics with automated mass photometry
- Architecture and Stoichiometry of Human Shelterin Using Single-Particle EM and Mass Analysis
- Studying protein polymerization with mass photometry
- Studying Form I Rubisco using ancestral sequence reconstruction and mass photometry
- Probing & modulating the function of the retromer endosomal trafficking complex
- Quantifying protein-protein interactions by molecular counting with mass photometry
- Using Mass Photometry to Quantitate CaMKII stoichiometries
- Brochures
-
- App notes
-
- Refeyn posts
-
- External Articles
-
Kofinova, Z., Karunanithy, G., Ferreira, A.S. and Struwe, W.B., 2024. Measuring protein‐protein interactions and quantifying their dissociation constants with mass photometry. Current Protocols, 4(1), p.e962 [Link]
Claasen, M., Kofinova, Z., Contino, M. and Struwe, W.B., 2024. Analysis of protein complex formation at micromolar concentrations by coupling microfluidics with mass Photometry. Journal of Visualized Experiments: Jove, (203). [Link]
Nagy, G.N., Zhao, X.F., Karlsson, R., Wang, K., Duman, R., Harlos, K., El Omari, K., Wagner, A., Clausen, H., Miller, R.L. and Giger, R.J., 2024. Structure and function of Semaphorin-5A glycosaminoglycan interactions. Nature Communications, 15(1), p.2723. [Link]
Awasthi, S., Ying, C., Li, J. and Mayer, M., 2023. Simultaneous determination of the size and shape of single α-synuclein oligomers in solution. ACS nano, 17(13), pp.12325-12335. [Link]
Wu, D. and Piszczek, G., 2020. Measuring the affinity of protein-protein interactions on a single-molecule level by mass photometry. Analytical biochemistry, 592, p.113575. [Link]
- Tau protein aggregation & neurodegenerative disease; new insights from mass photometry
- Characterizing protein oligomerization dynamics with automated mass photometry
- Architecture and Stoichiometry of Human Shelterin Using Single-Particle EM and Mass Analysis
- Studying protein polymerization with mass photometry
- Studying Form I Rubisco using ancestral sequence reconstruction and mass photometry
- Probing & modulating the function of the retromer endosomal trafficking complex
- Quantifying protein-protein interactions by molecular counting with mass photometry
- Using Mass Photometry to Quantitate CaMKII stoichiometries
