Mass photometry characterizes nucleic acids, proteins and their interactions
Interactions between nucleic acids and proteins are critical for many cellular processes, including DNA repair, replication and transcription. However, protein-nucleic acid interactions often result in highly heterogenous complexes, both in the type of binding partners and in the stoichiometry of the interactions. This heterogeneity presents a challenge to conventional bioanalytical methods.
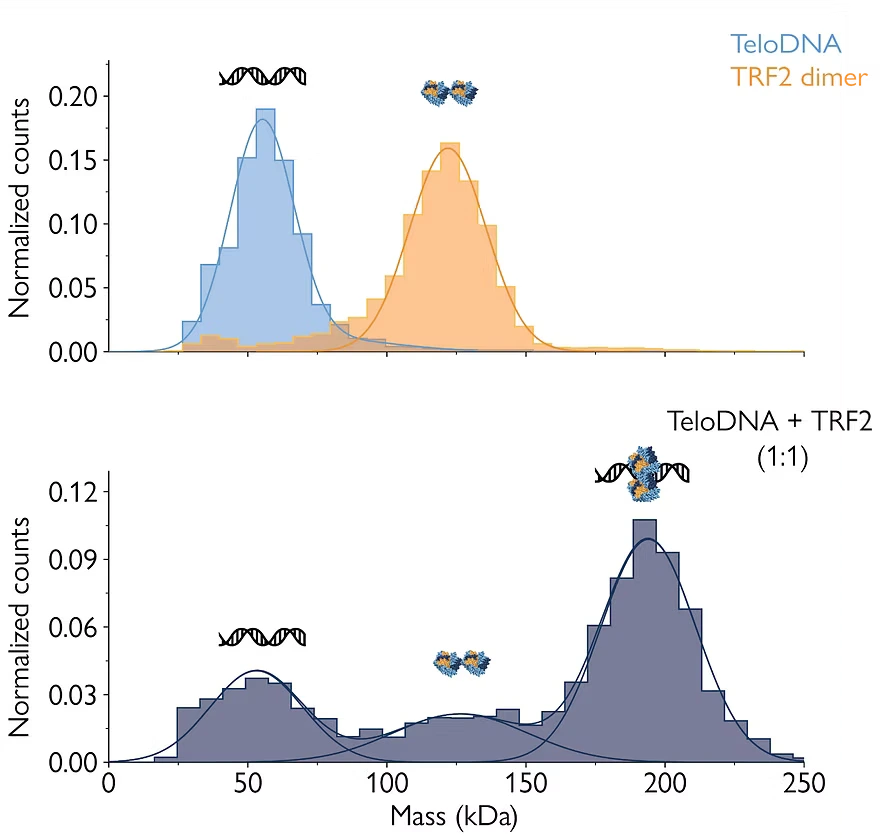
Mass photometry is a powerful technology that analyzes the mass distribution of the molecules present in a sample at the single-molecule level, without labels or immobilization.
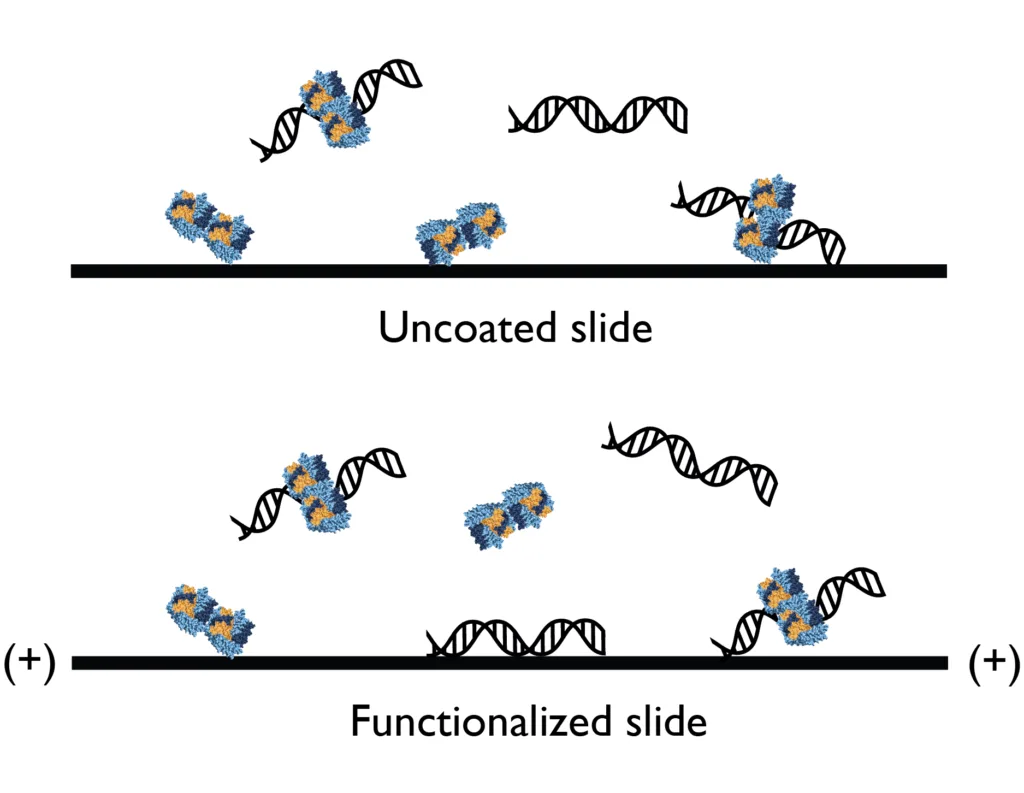
Mass photometry provides a detailed overview of all the molecules present in a sample as well as the complexes they form, even when they are present at low abundance. To analyze nucleic acids and their interactions with proteins with mass photometry, sample carrier slides need to be functionalized with poly-L-lysine (PLL) due to the nucleic acids’ negative
charge.
In combination with slides with a cationic coating, mass photometry can measure DNA, RNA, proteins and their interactions – no further adjustments needed! Get in touch with us for instructions on how to coat your sample carrier slides with PLL.
Measuring protein-DNA affinity with mass photometry vs. SPR
Mass photometry gives information about the proportions of biomolecular species in a sample – making it possible to quantify their binding affinities. As this application note demonstrates, dissociation constants (KD) for nucleic acid-protein complexes calculated from mass photometry measurements agree with results from surface plasmon resonance (SPR) – the analytical method considered the gold standard for assessing protein-DNA binding affinities.
To find out how to quickly quantify protein and DNA binding affinities
Characterizing protein-DNA interactions with mass photometry
Learn how mass photometry is a label-free bioanalytical tool that can characterize protein-DNA interactions by directly measuring the
relative abundance of proteins, nucleic acids and the complexes they form in solution. The use of Refeyn’s MassGlass® cationic
coating (CC) slides facilitates the study of protein-DNA interactions, since all binding partners can be characterized in a single
measurement – reducing experimental variability and saving users’ time.
Additional resources
WEBINAR: Elucidating the composition of CRISPR-Cas12f1 complexes using mass photometry
This webinar explores how the CRISPR-Cas12f1 effector complex recognizes and cleaves DNA. Mass photometry was used to measure the stoichiometry of two miniature Cas12f1 ribonucleoprotein complexes, AsCas12f1 and SpCas12f1 in vitro. The results indicate that the Cas protein forms a binary complex with guide RNA and remains bound when interacting with target DNA, forming a ternary complex. Overall, the results reveal the composition of a compact CRISPR-Cas system and its impact on target recognition and DNA cleavage mechanisms.


BLOG: Studying protein-DNA interactions with mass photometry
In this blog post, we present a recent study that used mass photometry to investigate protein-DNA interactions. We then speak with the paper’s first author, Ananya Acharya, who explains how she and her colleagues used mass photometry to answer unique scientific questions about DNA repair mechanisms and compares it to other techniques.
To learn more about how mass photometry can improve your bioanalytical processes
More Application Notes
Browse through our catalogue of application notes highlighting some recent case studies featuring mass photometry.
