Accelerate mRNA characterization with nanogram-scale sensitivity
mRNA vaccines and therapeutics have emerged as transformative medicines. However, conventional analytics are not designed for the speed and efficiency required in modern drug development and manufacturing. At Refeyn, we provide cutting-edge technology, using mass photometry, to give rapid, multiple attribute insights into intact mRNA of all sizes-using just nanograms of sample.
Key pain points experienced by researchers:
Resource-intensive analysis
Comprehensive analysis requires many analytical tests and a large volume of sample-making mRNA characterization time-consuming and resource-intensive.
Limited analytical capabilities
Legacy analytics struggle to accurately characterize longer and more complex mRNA molecules.
Loss of native structure
Most analytical techniques are denaturing, so they fail to capture true native behavior.
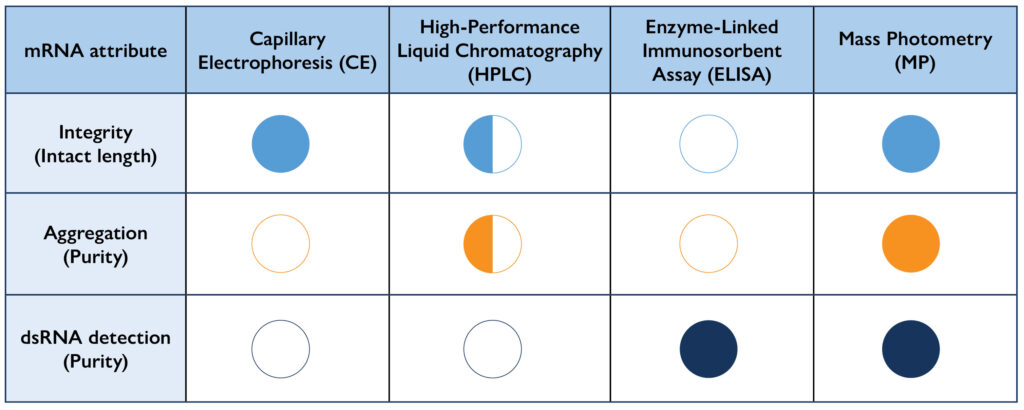
Figure 1. Comparison of Techniques used to measure critical quality attributes for mRNA. Mass photometry can measure multiple mRNA CQAs that are presently done by various techniques including CE, HPLC, and ELISA.
Challenges
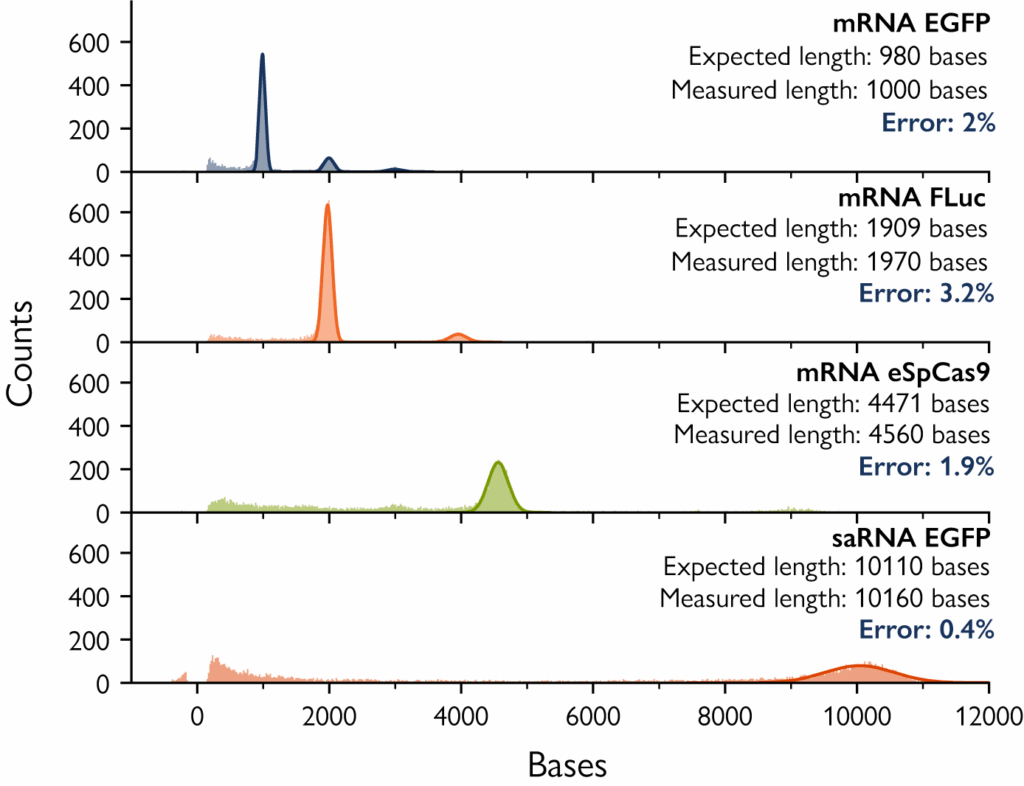
Figure 2. Mass photometry accurately measures mRNA and saRNA length. Samples of mRNA and saRNA, varying in length from 980 bases to 10.2 kb, were analyzed by mass photometry. In every case, the mass photometry measurement was highly accurate, returning lengths in close agreement with the expected values.
Accurate measurements of long mRNAs
Accurately measuring the length of long mRNAs, such as those encoding Cas9 (>4 kb), is a persistent challenge in therapeutic development. These large transcripts are prone to partial degradation, and even small deviations in length can reduce translation efficiency, alter protein folding, or impact potency. From early discovery to manufacturing scale-up, confirming that each long mRNA is full-length and intact is essential for ensuring product quality, regulatory compliance, and therapeutic efficacy.
Mass photometry delivers a unique solution by quickly measuring long mRNA length and integrity, with <5% relative error and no labels or sample preparation. By directly detecting individual molecules in solution, mass photometry provides the resolution needed to detect subtle truncations or degradation products, which are often missed by conventional sizing techniques.
In contrast, capillary gel electrophoresis (CGE) requires lengthy workflows, can introduce artifacts from denaturation or labeling, and typically achieves only 15–30% relative error-making it less reliable for high-precision analysis of large, therapeutically relevant mRNAs.
Native-state mRNA measurements
mRNA integrity is a critical determinant of therapeutic safety and quality. Even minor fragmentation can shorten half-life, reduce protein yield, or trigger immune responses so detecting integrity is essential. Degradation studies are routinely used to assess stability under stress conditions, making it essential to quickly assess samples.
Mass photometry offers direct visualization of intact and degraded mRNA in native conditions, delivering results in minutes with <5% relative error. Its high precision allows detection of subtle integrity changes across time points or stress cycles without artifacts introduced by denaturation or labeling. Unlike CGE, mass photometry preserves true degradation profiles and requires minimal sample. Mass photometry’s speed and sensitivity enable rapid, data-driven decisions on stability and process robustness.
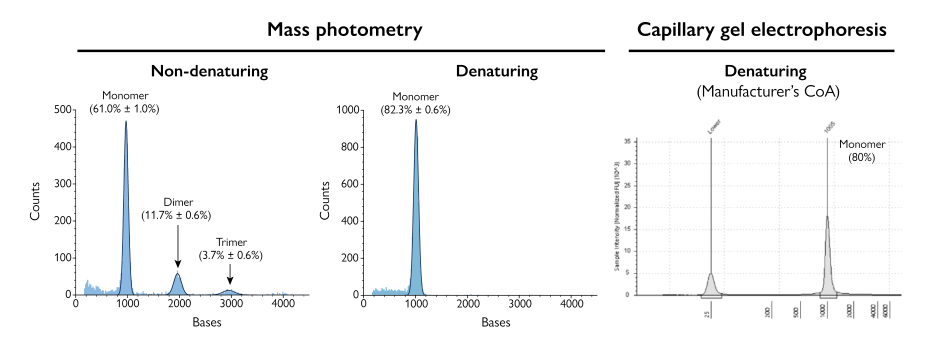
Figure 3. Mass photometry does not require denaturation, so it can detect species that are missed by other techniques. Shown here are measurements of eGFP mRNA. The mass photometry results in denaturing conditions align with those from CGE, but the mass photometry result under non-denaturing conditions detects additional molecular species (e.g., monomer, dimer, trimer).
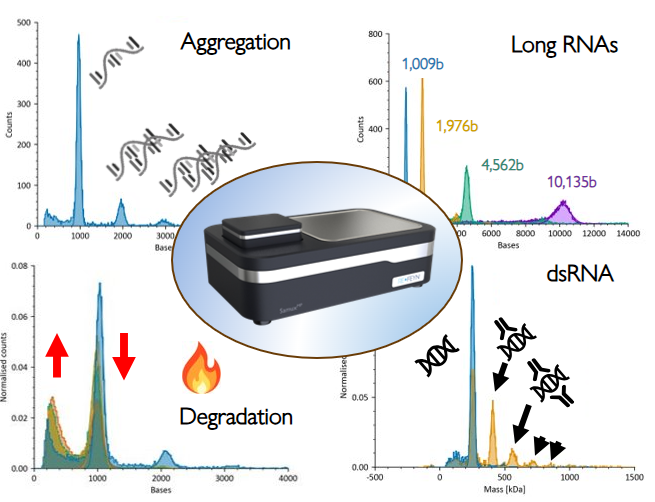
Figure 4. One assay, multiple attributes. A single mass photometry measurement can report on multiple mRNA CQAs, such as:. mRNA aggregation under native conditions, long RNA length (from ~1,000 to over 10,000 bases), degradation profiles showing loss of intact transcript and accumulation of shorter fragments, and quantification of dsRNA impurities alongside the main mRNA population. This unified, label-free approach allows rapid, high-resolution assessment of integrity, purity, size, and secondary structure-related species without the need for multiple orthogonal assays.
Assessing multiple attributes
Analyzing mRNA therapeutics is complex. Traditional methods require multiple assays to assess transcript length, detect impurities, and monitor degradation, often involving dyes, labels, or denaturing conditions that can distort results. These fragmented workflows slow down decision-making and increase the risk of artifacts.
Mass photometry offers a streamlined solution. It enables label-free, single-molecule analysis in native buffers using just nanograms of sample. With base-level precision, it measures mRNA length, detects aggregation and higher order complexes, quantifies double-stranded RNA impurities, and tracks degradation products all on one platform.
With just a single rapid measurement, mass photometry provides accurate data on the purity and integrity of your product and reflects its true molecular state. Whether verifying long transcripts like Cas9, assessing freeze/thaw stability, or ensuring regulatory compliance, mass photometry empowers scientists to make confident, data-driven decisions across discovery, development, and manufacturing.
Mass photometry solutions for mRNA characterization
Refeyn TwoMP & TwoMP Auto
The TwoMP is a powerful mass photometer engineered for rapid, label-free characterization of mRNA therapeutics at the single-molecule level.

It provides unique insights into individual mRNA molecules by measuring their mass in native buffer conditions, eliminating the need for labels or extensive sample preparation. This makes it an essential tool for research, development, and quality control in mRNA-based therapies.
Its core features address key analytical challenges in mRNA characterization:
Minimal Sample Requirements
Requires only nanograms of sample for analysis, preserving precious material.
Rapid Analysis
Delivers comprehensive mRNA characterization data within minutes per sample.
Native Single-Molecule Precision
Utilizes single-molecule resolution to accurately measure molecular mass under native conditions.
MassGlass™ NA Slides
A high-quality surface that promotes efficient mRNA binding is essential for mass photometry measurements.
MassGlass™ NA Slides provide a ready-to-use, out-of-the-box solution that consistently delivers reliable performance in mRNA mass photometry measurements. Their advanced surface technology ensures strong mRNA binding, enabling accurate quantification analysis.
Refeyn offers MassGlass™ NA – Sample Prep Kits for the TwoMP and TwoMP Auto systems, containing everything you need for a seamless experience when measuring your mRNA samples.
Purpose-built to tackle the complexities of mRNA analysis with precision and efficiency, MassGlass™ NA Slides offer:
A Plug-and-Play Solution
Streamlines mRNA analysis with an out-of-the-box solution.
Guaranteed Quality
Ensures a high quality mRNA measurement every time.
Extended Shelf Life
Delivers consistent performance for up to four weeks after opening the box, ensuring extended usability and reliability.
AcquireMP acquisition and DiscoverMP analysis software NA
Refeyn’s AquireMP acquisition and DiscoverMP analysis software provides an intuitive, high-performance solution for single-molecule mass photometry workflows.
Designed to streamline both data capture and interpretation, the software suite enables real-time measurement, clear user guidance, and powerful analysis tools–making it an essential companion for researchers and analysts working with complex biomolecular samples.
Its core features support fast, confident decision-making:
User-Friendly Interface
Intuitive interface with minimal parameters to optimize.
Real-Time Quality Control
Real-time results and quality metrics ensure high-quality data capture.
Instant Report Generation
Advanced analysis and graphing tools deliver report-ready insights in minutes.

- Handbook
mRNA characterization handbook
Discover how our cutting-edge mass photometry technology delivers fast, native mRNA analysis for multiple attributes and across a broad range of mRNA sizes.
Mass photometry in nucleic acids literature
Peer-reviewed evidence for mass photometry
Mass photometry's “Matrix robustness and overall simplicity of sampling is a large boon for in-process sampling” Schmudlach, et al. (2025). Biology Methods and Protocols 10, bpaf021.
https://doi.org/10.1093/biomethods/bpaf021
"Mass photometry has an advantage over CDMS of reflecting in an unbiased manner the presence and amounts of both LMWS and HMWS in a sample.” Desligniere et al (2025) Mol Ther Methods Clin Dev, 33, 101454.
https://doi.org/10.1016/j.omtm.2025.101454
"Consistent measurements of mRNA lengths were observed with calculated errors of less than 2.3%… demonstrating the robustness and accuracy of MP as a reliable tool for accurately determining mRNA length across a wide range of sizes.” Camperi et al. (2024). Anal. Chem., 96, 3886–3897.
https://pubs.acs.org/doi/10.1021/acs.analchem.3c05539
Resources

Accelerate mRNA Analytics with Mass Photometry
Best practices for mass photometry analysis of mRNA: Unlike proteins, mRNA’s conformational flexibility and ion sensitivity demand extra care when using mass photometry. This white paper dives into how experimental conditions can make or break your measurements — and offers practical tips to optimize your setup for clean, reproducible results.

Assessing mRNA stability and integrity using mass photometry
Discover how mass photometry is reshaping mRNA-based therapeutics. This high-level webinar offers a deep dive into published applications and analytical strategies for characterizing mRNA, including mRNA vaccines.
Method development for characterization of mRNA vaccines and therapeutics with mass photometry
Learn how mRNA is transforming therapeutics and see how mass photometry boosts data quality, workflow efficiency, and QC.

Rapid, quantitative, multi-attribute mass photometry analysis for mRNA stability studies
Mass photometry (MP) enables rapid, high-resolution analysis of mRNA integrity and purity under native conditions. It provides single-molecule, quantitative measurements of full-length transcripts, degraded fragments, and aggregates within minutes.
Frequently asked questions
How long does a mass photometry experiment take to measure mRNA?
A mass photometry experiment can be prepared, collected, and analyzed in under 5 minutes.
What is the range of mRNA lengths that can be measured by mass photometry?
We have successfully measured RNA samples between 200 and 10,000 bases with high accuracy (<5% error).
What sample concentration of mRNA is required for reliable detection in mass photometry?
Each measurement requires 10-20 µL of sample with a concentrations in the range of 1-10 nM (roughly 10–100 ng/µL for a 1 kb mRNA). Lower concentrations may work with a highly purified sample (i.e., a sample with very low background noise).
What buffers are compatible with mass photometry measurements of mRNA?
Mass photometry is generally buffer-insensitive within moderate ionic strength. Common choices: 10 mM HEPES or Tris, pH 7–8, with up to 150 mM NaCl. Avoid high glycerol or viscous additives.
Can I use MassGlass UC for RNA measurements?
No, the strong negative charge of RNA requires a cationic coating on the slide. MassGlass™ NA has been optimized for capture, resolution, reproducibility, and sample integrity. Order MassGlass™ NA today
Is the surface charge of RNA important for binding to the slide surface?
Yes, surface charge is important for the macromolecules’ ability to bind to the slide surface, which is necessary for mass photometry measurements. While Refeyn’s bare glass MassGlass UC slides are perfectly suitable for analyzing proteins with mass photometry, RNA has a large negative charge from the phosphate backbone and will not bind directly to negatively charged glass (silica) slides (MassGlass UC slides). A positively charged coating is necessary on the slide’s surface. Refeyn’s MassGlass NA slides, specifically designed for measuring mRNA samples, have the necessary coating.
What standards should be used to calibrate mass photometry measurements of RNA?
Refeyn recommends using ssRNA ladders (e.g. NEB N0364; ThermoFisher SM1821) for calibration. This is also what authors of scientific publications that use mass photometry to analyze mRNA have done.
How much sample preparation is required for measuring mRNA with mass photometry?
In most cases, only a simple dilution step is required. Mass photometry can measure samples in a wide variety of buffer conditions in the range of 1-10 nM mRNA. No denaturation or specialized reagents are required, which allows investigation of the native and oligomeric distribution of the mRNA sample.
How accurate is mass photometry at measuring mRNA length?
We have qualified <5% relative error for measured lengths of mRNAs up to 10 kb using mass photometry. By comparison, orthogonal techniques like Capillary Electrophoresis are typically benchmarked with 15-30% relative error on mRNA length measurements.
Is it possible to quantify mRNA purity using mass photometry?
Yes, mass photometry quickly measures the proportion of full-length mRNA versus fragments or aggregates under native conditions, and with only nanograms of sample.
Can I measure siRNA with mass photometry?
siRNA are generally below the limit of detection for mass photometry, which is around 200 bases.
How does the mass photometry signal differ for mRNA versus proteins?
RNA has more structural flexibility than proteins, which results in broader mass peaks. The full width at half maximum (FWHM) for RNA mass peaks is typically ~5-10%, compared to ~2% for proteins.
Can mass photometry be used to study mRNA–protein interactions (e.g., RNP complexes)?
Yes. Binding of proteins to mRNA can be measured and should be reported as mass shifts relative to the pure species. Experiments with ribonucleoproteins and Cas9/sgRNA complexes demonstrate this principle (Bigelyte et al, 2021 Nat. Comm.12, 6191).
Unlock faster mRNA insights with
mass photometry
Let's get started! If you have any other questions or would like further details, please get in touch!

Accelerate mRNA Analytics with Mass Photometry
Best practices for mass photometry analysis of mRNA: Unlike proteins, mRNA’s conformational flexibility and ion sensitivity demand extra care when using mass photometry. This white paper dives into how experimental conditions can make or break your measurements — and offers practical tips to optimize your setup for clean, reproducible results.

Assessing mRNA stability and integrity using mass photometry
Discover how mass photometry is reshaping mRNA-based therapeutics. This high-level webinar offers a deep dive into published applications and analytical strategies for characterizing mRNA, including mRNA vaccines.

Method development for characterization of mRNA vaccines and therapeutics with mass photometry
Learn how mRNA is transforming therapeutics and see how mass photometry boosts data quality, workflow efficiency, and QC.